Takes slices of the spatiotemporal kernel density or relative risk function estimate at desired times
spattemp.slice(stob, tt, checkargs = TRUE)Arguments
- stob
An object of class
stdenorrrstgiving the spatiotemporal estimate from which to take slices.- tt
Desired time(s); the density/risk surface estimate corresponding to which will be returned. This value must be in the available range provided by
stob$tlim; see `Details'.- checkargs
Logical value indicating whether to check validity of
stobandtt. Disable only if you know this check will be unnecessary.
Value
A list of lists of pixel images, each of which corresponds to
the requested times in tt, and are named as such.
If stob is an object of class stden:
- z
Pixel images of the joint spatiotemporal density corresponding to
tt.- z.cond
Pixel images of the conditional spatiotemporal density given each time in
tt.
If stob is an object of class rrst:
- rr
Pixel images of the joint spatiotemporal relative risk corresponding to
tt.- rr.cond
Pixel images of the conditional spatiotemporal relative risk given each time in
tt.- P
Only present if
tolerate = TRUEin the preceding call tospattemp.risk. Pixel images of the \(p\)-value surfaces for the joint spatiotemporal relative risk.- P.cond
Only present if
tolerate = TRUEin the preceding call tospattemp.risk. Pixel images of the \(p\)-value surfaces for the conditional spatiotemporal relative risk.
Details
Contents of the stob argument are returned based on a discretised set of times.
This function internally computes the desired surfaces as
pixel-by-pixel linear interpolations using the two discretised times
that bound each requested tt.
The function returns an error if any of the
requested slices at tt are not within the available range of
times as given by the tlim
component of stob.
References
Fernando, W.T.P.S. and Hazelton, M.L. (2014), Generalizing the spatial relative risk function, Spatial and Spatio-temporal Epidemiology, 8, 1-10.
See also
Examples
# \donttest{
data(fmd)
fmdcas <- fmd$cases
fmdcon <- fmd$controls
f <- spattemp.density(fmdcas,h=6,lambda=8)
#> Calculating trivariate smooth...
#> Done.
#> Edge-correcting...
#> Done.
#> Conditioning on time...
#> Done.
g <- bivariate.density(fmdcon,h0=6)
rho <- spattemp.risk(f,g,tolerate=TRUE)
#> Calculating ratio...
#> Done.
#> Ensuring finiteness...
#> --joint--
#> --conditional--
#> Done.
#> Calculating tolerance contours...
#> --convolution 1--
#> --convolution 2--
#> Done.
f$tlim # requested slices must be in this range
#> [1] 20 220
# slicing 'stden' object
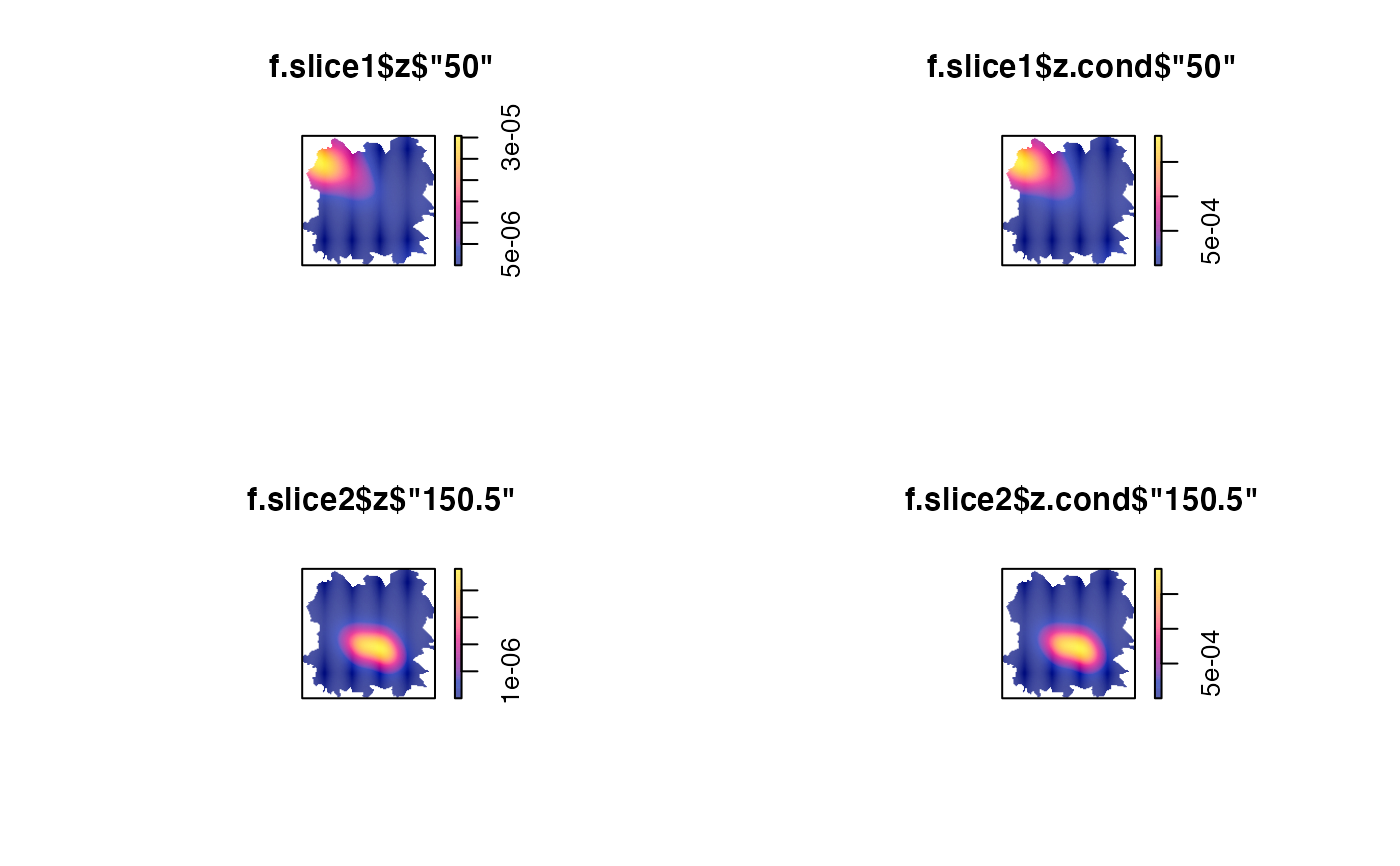
f.slice1 <- spattemp.slice(f,tt=50) # evaluation timestamp
f.slice2 <- spattemp.slice(f,tt=150.5) # interpolated timestamp
par(mfrow=c(2,2))
plot(f.slice1$z$'50')
plot(f.slice1$z.cond$'50')
plot(f.slice2$z$'150.5')
plot(f.slice2$z.cond$'150.5')
 # slicing 'rrst' object
rho.slices <- spattemp.slice(rho,tt=c(50,150.5))
par(mfrow=c(2,2))
plot(rho.slices$rr$'50');tol.contour(rho.slices$P$'50',levels=0.05,add=TRUE)
plot(rho.slices$rr$'150.5');tol.contour(rho.slices$P$'150.5',levels=0.05,add=TRUE)
plot(rho.slices$rr.cond$'50');tol.contour(rho.slices$P.cond$'50',levels=0.05,add=TRUE)
plot(rho.slices$rr.cond$'150.5');tol.contour(rho.slices$P.cond$'150.5',levels=0.05,add=TRUE)
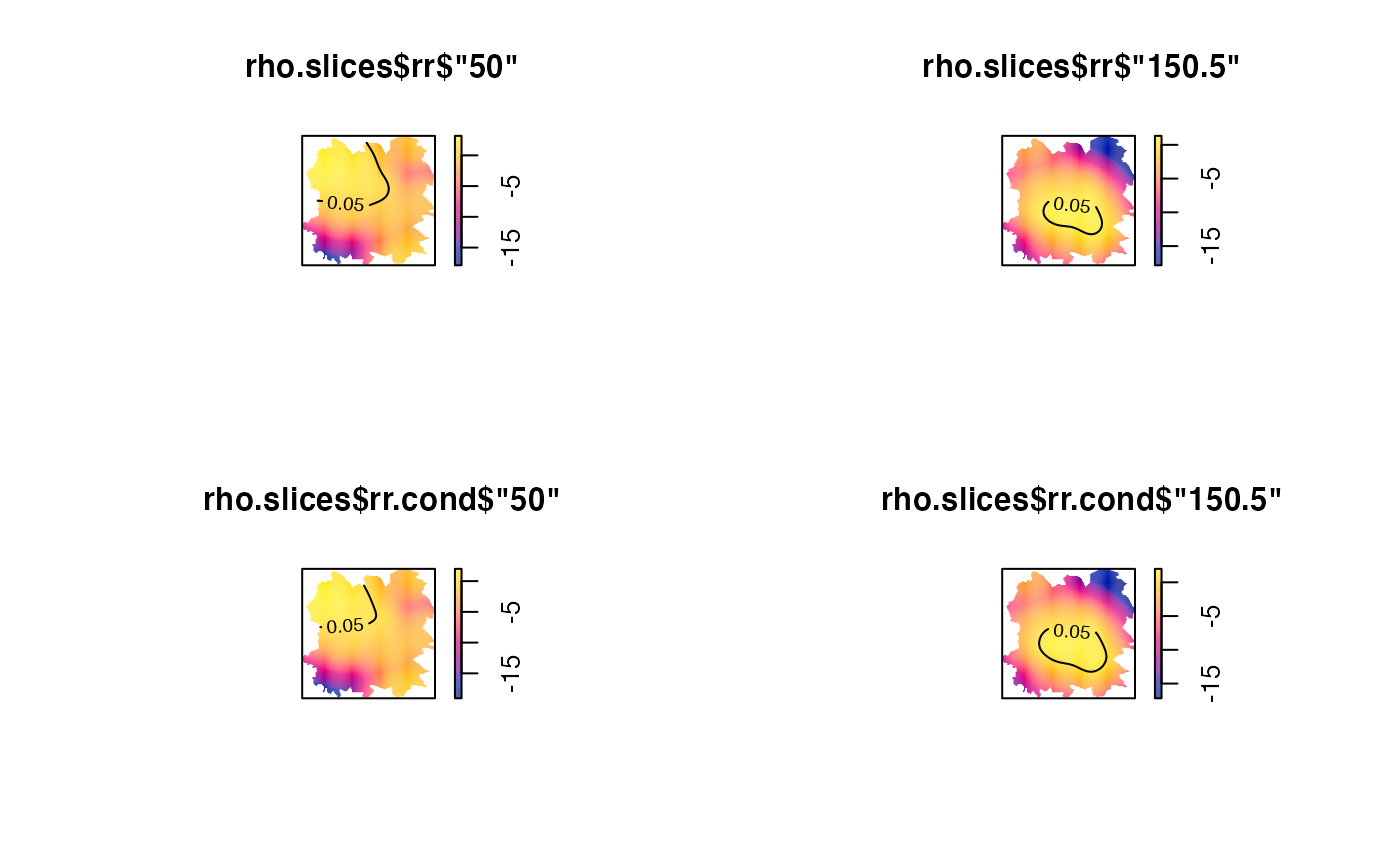
# slicing 'rrst' object
rho.slices <- spattemp.slice(rho,tt=c(50,150.5))
par(mfrow=c(2,2))
plot(rho.slices$rr$'50');tol.contour(rho.slices$P$'50',levels=0.05,add=TRUE)
plot(rho.slices$rr$'150.5');tol.contour(rho.slices$P$'150.5',levels=0.05,add=TRUE)
plot(rho.slices$rr.cond$'50');tol.contour(rho.slices$P.cond$'50',levels=0.05,add=TRUE)
plot(rho.slices$rr.cond$'150.5');tol.contour(rho.slices$P.cond$'150.5',levels=0.05,add=TRUE)
 # }
# }