Provides an isotropic adaptive or fixed bandwidth kernel density/intensity estimate of bivariate/planar/2D data.
bivariate.density(
pp,
h0,
hp = NULL,
adapt = FALSE,
resolution = 128,
gamma.scale = "geometric",
edge = c("uniform", "diggle", "none"),
weights = NULL,
intensity = FALSE,
trim = 5,
xy = NULL,
pilot.density = NULL,
leaveoneout = FALSE,
parallelise = NULL,
davies.baddeley = NULL,
verbose = TRUE
)Arguments
- pp
An object of class
pppgiving the observed 2D data set to be smoothed.- h0
Global bandwidth for adaptive smoothing or fixed bandwidth for constant smoothing. A numeric value > 0.
- hp
Pilot bandwidth (scalar, numeric > 0) to be used for fixed bandwidth estimation of a pilot density in the case of adaptive smoothing. If
NULL(default), it will take on the value ofh0. Ignored whenadapt = FALSEor ifpilot.densityis supplied as a pre-defined pixel image.- adapt
Logical value indicating whether to perform adaptive kernel estimation. See `Details'.
- resolution
Numeric value > 0. Resolution of evaluation grid; the density/intensity will be returned on a [
resolution\(\times\)resolution] grid.- gamma.scale
Scalar, numeric value > 0; controls rescaling of the variable bandwidths. Defaults to the geometric mean of the bandwidth factors given the pilot density (as per Silverman, 1986). See `Details'.
- edge
Character string giving the type of edge correction to employ.
"uniform"(default) corrects based on evaluation grid coordinate and"diggle"reweights each observation-specific kernel. Settingedge = "none"requests no edge correction. Further details can be found in the documentation fordensity.ppp.- weights
Optional numeric vector of nonnegative weights corresponding to each observation in
pp. Must have length equal tonpoints(pp).- intensity
Logical value indicating whether to return an intensity estimate (integrates to the sample size over the study region), or a density estimate (default, integrates to 1).
- trim
Numeric value > 0; controls bandwidth truncation for adaptive estimation. See `Details'.
- xy
Optional alternative specification of the evaluation grid; matches the argument of the same tag in
as.mask. If supplied,resolutionis ignored.- pilot.density
An optional pixel image (class
im) giving the pilot density to be used for calculation of the variable bandwidths in adaptive estimation, or appp.objectgiving the data upon which to base a fixed-bandwidth pilot estimate usinghp. If used, the pixel image must be defined over the same domain as the data givenresolutionor the supplied pre-setxyevaluation grid; or the planar point pattern data must be defined with respect to the same polygonal study region as inpp.- leaveoneout
Logical value indicating whether to compute and return the value of the density/intensity at each data point for an adaptive estimate. See `Details'.
- parallelise
Numeric argument to invoke parallel processing, giving the number of CPU cores to use when
leaveoneout = TRUE. Experimental. Test your system first usingparallel::detectCores()to identify the number of cores available to you.- davies.baddeley
An optional numeric vector of length 3 to control bandwidth partitioning for approximate adaptive estimation, giving the quantile step values for the variable bandwidths for density/intensity and edge correction surfaces and the resolution of the edge correction surface. May also be provided as a single numeric value. See `Details'.
- verbose
Logical value indicating whether to print a function progress bar to the console when
adapt = TRUE.
Value
If leaveoneout = FALSE, an object of class "bivden".
This is effectively a list with the following components:
- z
The resulting density/intensity estimate, a pixel image object of class
im.- h0
A copy of the value of
h0used.- hp
A copy of the value of
hpused.- h
A numeric vector of length equal to the number of data points, giving the bandwidth used for the corresponding observation in
pp.- him
A pixel image (class
im), giving the `hypothetical' Abramson bandwidth at each pixel coordinate conditional upon the observed data.NULLfor fixed-bandwidth estimates.- q
Edge-correction weights; a pixel
image ifedge = "uniform", a numeric vector ifedge = "diggle", andNULLifedge = "none".- gamma
The value of \(\gamma\) used in scaling the bandwidths.
NAif a fixed bandwidth estimate is computed.- geometric
The geometric mean \(G\) of the untrimmed bandwidth factors \(\tilde{f}(x_i)^{-1/2}\).
NAif a fixed bandwidth estimate is computed.- pp
A copy of the
ppp.objectinitially passed to theppargument, containing the data that were smoothed.
Else, if leaveoneout = TRUE, simply a numeric vector of length equal to the
number of data points, giving the leave-one-out value of the function at the
corresponding coordinate.
Details
Given a data set \(x_1,\dots,x_n\) in 2D, the isotropic kernel estimate of its probability density function, \(\hat{f}(x)\), is given by $$\hat{f}(y)=n^{-1}\sum_{i=1}^{n}h(x_i)^{-2}K((y-x_i)/h(x_i)) $$ where \(h(x)\) is the bandwidth function, and \(K(.)\) is the bivariate standard normal smoothing kernel. Edge-correction factors (not shown above) are also implemented.
- Fixed
The classic fixed bandwidth kernel estimator is used when
adapt = FALSE. This amounts to setting \(h(u)=\)h0for all \(u\). Further details can be found in the documentation fordensity.ppp.- Adaptive
Setting
adapt = TRUErequests computation of Abramson's (1982) variable-bandwidth estimator. Under this framework, we have \(h(u)=\)h0*min[\(\tilde{f}(u)^{-1/2}\),\(G*\)trim]/\(\gamma\), where \(\tilde{f}(u)\) is a fixed-bandwidth kernel density estimate computed using the pilot bandwidthhp.Global smoothing of the variable bandwidths is controlled with the global bandwidth
h0.In the above statement, \(G\) is the geometric mean of the ``bandwidth factors'' \(\tilde{f}(x_i)^{-1/2}\); \(i=1,\dots,n\). By default, the variable bandwidths are rescaled by \(\gamma=G\), which is set with
gamma.scale = "geometric". This allowsh0to be considered on the same scale as the smoothing parameter in a fixed-bandwidth estimate i.e. on the scale of the recorded data. You can use any other rescaling ofh0by settinggamma.scaleto be any scalar positive numeric value; though note this only affects \(\gamma\) -- see the next bullet. When using a scale-invarianth0, setgamma.scale = 1.The variable bandwidths must be trimmed to prevent excessive values (Hall and Marron, 1988). This is achieved through
trim, as can be seen in the equation for \(h(u)\) above. The trimming of the variable bandwidths is universally enforced by the geometric mean of the bandwidth factors \(G\) independent of the choice of \(\gamma\). By default, the function truncates bandwidth factors at five times their geometric mean. For stricter trimming, reducetrim, for no trimming, settrim = Inf.For even moderately sized data sets and evaluation grid
resolution, adaptive kernel estimation can be rather computationally expensive. The argumentdavies.baddeleyis used to approximate an adaptive kernel estimate by a sum of fixed bandwidth estimates operating on appropriate subsets ofpp. These subsets are defined by ``bandwidth bins'', which themselves are delineated by a quantile step value \(0<\delta<1\). E.g. setting \(\delta=0.05\) will create 20 bandwidth bins based on the 0.05th quantiles of the Abramson variable bandwidths. Adaptive edge-correction also utilises the partitioning, with pixel-wise bandwidth bins defined using the value \(0<\beta<1\), and the option to decrease the resolution of the edge-correction surface for computation to a [\(L\) \(\times\) \(L\)] grid, where \(0 <L \le\)resolution. Ifdavies.baddeleyis supplied as a vector of length 3, then the values[1], [2], and [3]correspond to the parameters \(\delta\), \(\beta\), and \(L_M=L_N\) in Davies and Baddeley (2018). If the argument is simply a single numeric value, it is used for both \(\delta\) and \(\beta\), with \(L_M=L_N=\)resolution(i.e. no edge-correction surface coarsening).Computation of leave-one-out values (when
leaveoneout = TRUE) is done by brute force, and is therefore very computationally expensive for adaptive smoothing. This is because the leave-one-out mechanism is applied to both the pilot estimation and the final estimation stages. Experimental code to do this via parallel processing using theforeachroutine is implemented. Fixed-bandwidth leave-one-out can be performed directly indensity.ppp.
References
Abramson, I. (1982). On bandwidth variation in kernel estimates --- a square root law, Annals of Statistics, 10(4), 1217-1223.
Davies, T.M. and Baddeley A. (2018), Fast computation of spatially adaptive kernel estimates, Statistics and Computing, 28(4), 937-956.
Davies, T.M. and Hazelton, M.L. (2010), Adaptive kernel estimation of spatial relative risk, Statistics in Medicine, 29(23) 2423-2437.
Davies, T.M., Jones, K. and Hazelton, M.L. (2016), Symmetric adaptive smoothing regimens for estimation of the spatial relative risk function, Computational Statistics & Data Analysis, 101, 12-28.
Diggle, P.J. (1985), A kernel method for smoothing point process data, Journal of the Royal Statistical Society, Series C, 34(2), 138-147.
Hall P. and Marron J.S. (1988) Variable window width kernel density estimates of probability densities. Probability Theory and Related Fields, 80, 37-49.
Marshall, J.C. and Hazelton, M.L. (2010) Boundary kernels for adaptive density estimators on regions with irregular boundaries, Journal of Multivariate Analysis, 101, 949-963.
Silverman, B.W. (1986), Density Estimation for Statistics and Data Analysis, Chapman & Hall, New York.
Wand, M.P. and Jones, C.M., 1995. Kernel Smoothing, Chapman & Hall, London.
Examples
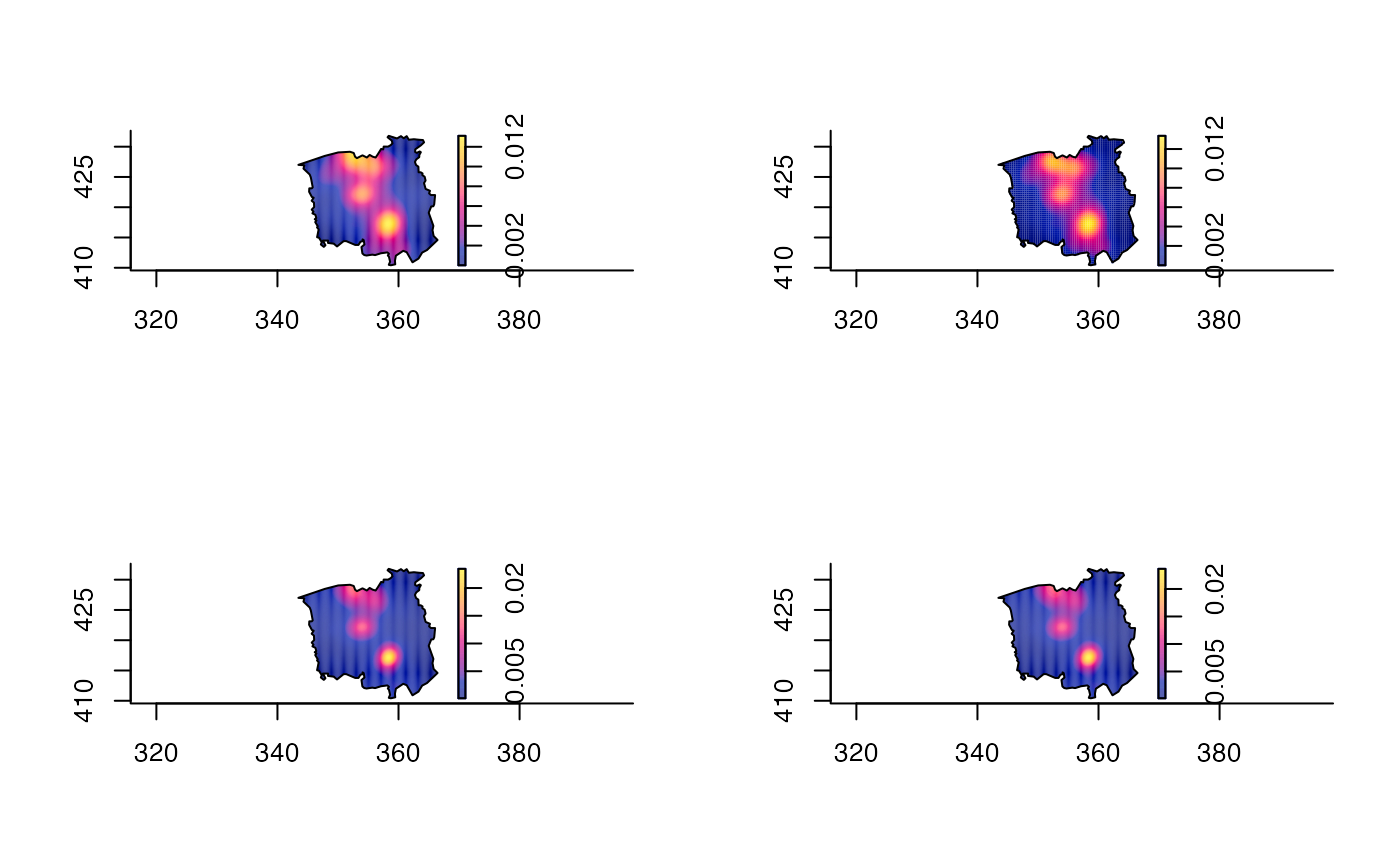
data(chorley) # Chorley-Ribble data from package 'spatstat'
# Fixed bandwidth kernel density; uniform edge correction
chden1 <- bivariate.density(chorley,h0=1.5)
# Fixed bandwidth kernel density; diggle edge correction; coarser resolution
chden2 <- bivariate.density(chorley,h0=1.5,edge="diggle",resolution=64)
# \donttest{
# Adaptive smoothing; uniform edge correction
chden3 <- bivariate.density(chorley,h0=1.5,hp=1,adapt=TRUE)
#> ================================================================================
# Adaptive smoothing; uniform edge correction; partitioning approximation
chden4 <- bivariate.density(chorley,h0=1.5,hp=1,adapt=TRUE,davies.baddeley=0.025)
#> ================================================================================
par(mfrow=c(2,2))
plot(chden1);plot(chden2);plot(chden3);plot(chden4)
 # }
# }